Matrix: Dehghani/light_in_tissue
Description: Light transport in soft tissue. Hamid Dehghani, Univ. Exeter, UK
 |
| (undirected graph drawing) |
 |
| Matrix properties | |
| number of rows | 29,282 |
| number of columns | 29,282 |
| nonzeros | 406,084 |
| structural full rank? | yes |
| structural rank | 29,282 |
| # of blocks from dmperm | 1 |
| # strongly connected comp. | 1 |
| explicit zero entries | 0 |
| nonzero pattern symmetry | symmetric |
| numeric value symmetry | 0% |
| type | complex |
| structure | unsymmetric |
| Cholesky candidate? | no |
| positive definite? | no |
| author | H. Dehghani |
| editor | T. Davis |
| date | 2007 |
| kind | electromagnetics problem |
| 2D/3D problem? | yes |
| Additional fields | size and type |
| b | sparse 29282-by-1 |
| Q | sparse 14641-by-1 |
| nodes | full 14641-by-3 |
| elements | full 28800-by-3 |
Notes:
% The problem is solving the fluence (PHI) of light in soft tissue using
% a simplified 3rd spherical harmonic expansion (SPN3) of the Radiative
% Transport Equation. There are two coupled equations to solve:
% M1*phi1 = Q + (M2*phi2) eq(1)
% (M4 - (M3*inv(M1)*M2))*phi2 = -2/3*Q + M3*inv(M1)*Q eq(2)
% PHI = phi1 - (1/3).*phi2 eq(3)
Problem = UFget ('Dehghani/light_in_tissue') ;
A = Problem.A ; % get the problem
Q = Problem.aux.Q ;
k = size (A,1) / 2 ;
M1 = A (1:k,1:k) ;
M2 = A (1:k,k+1:end) ;
M3 = A (k+1:end, 1:k) ;
M4 = A (k+1:end, k+1:end) ;
elements = Problem.aux.elements ;
nodes = Problem.aux.nodes ;
Q2 = (-(2/3).*Q) + (M3*(M1\Q)) ; % create rhs for equation 2
Q2 = [sparse(k,1) ; Q2] ;
phi2 = A\Q2 ; % solve for phi2
phi2 = phi2 (end/2+1:end,:) ;
Q1 = Q + M2*phi2 ; % calculate rhs for equation 1
phi1 = M1\Q1; % solve for phi1
PHI = phi1 - (1/3).*phi2;
figure (1) ; clf % plot results
trisurf(elements, nodes(:,1), nodes(:,2), nodes(:,3), log(abs(PHI))) ;
shading interp ;
view (2) ;
colorbar('horiz') ;
axis equal ;
axis off ;
colormap hot ;
| Ordering statistics: | result |
| nnz(chol(P*(A+A'+s*I)*P')) with AMD | 1,390,043 |
| Cholesky flop count | 2.0e+08 |
| nnz(L+U), no partial pivoting, with AMD | 2,750,804 |
| nnz(V) for QR, upper bound nnz(L) for LU, with COLAMD | 2,925,391 |
| nnz(R) for QR, upper bound nnz(U) for LU, with COLAMD | 5,672,099 |
| SVD-based statistics: | |
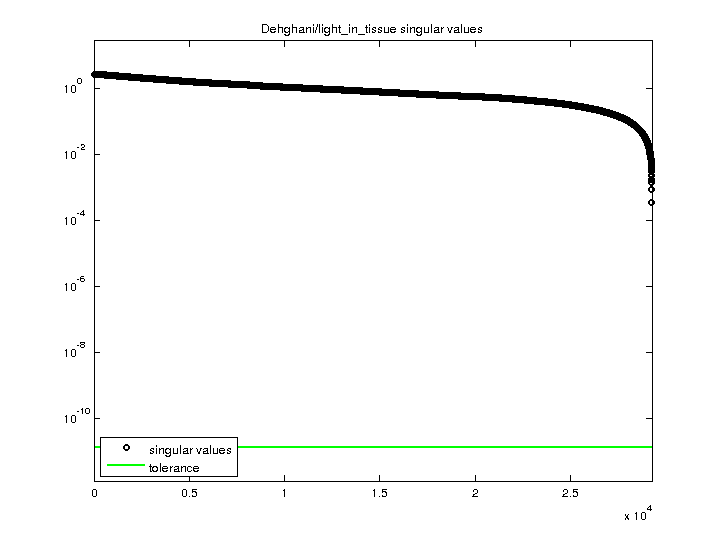
| norm(A) | 2.66355 |
| min(svd(A)) | 0.000340525 |
| cond(A) | 7821.9 |
| rank(A) | 29,282 |
| sprank(A)-rank(A) | 0 |
| null space dimension | 0 |
| full numerical rank? | yes |
| singular values (MAT file): | click here |
| SVD method used: | s = svd (full (A)) |
| status: | ok |
For a description of the statistics displayed above, click here.
Maintained by Tim Davis, last updated 12-Mar-2014.
Matrix pictures by cspy, a MATLAB function in the CSparse package.
Matrix graphs by Yifan Hu, AT&T Labs Visualization Group.